National
Science Foundation grants will bring together what's known about how species
are related
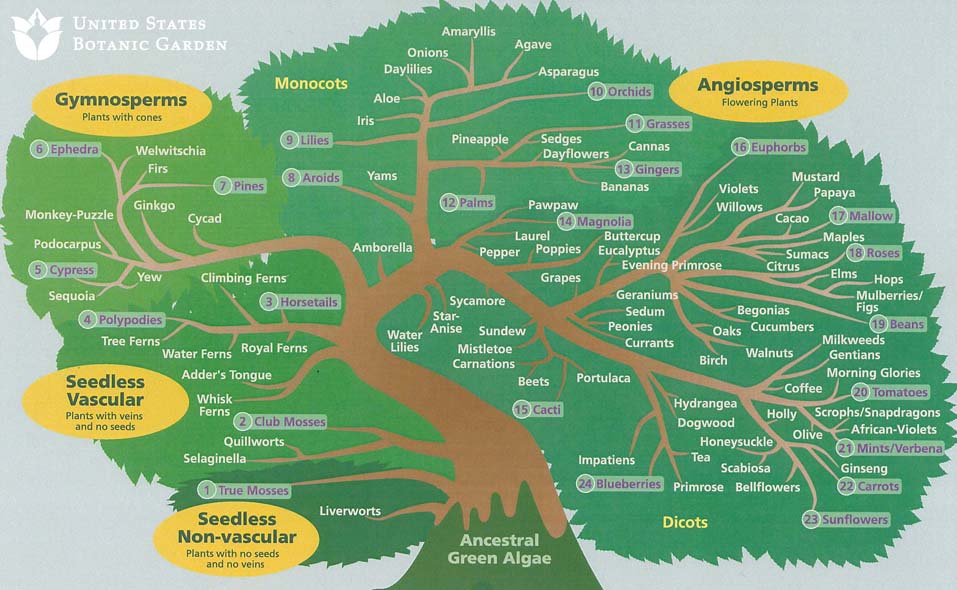
 A new initiative aims to build a
comprehensive tree of life that brings together everything scientists know
about how all species are related, from the tiniest bacteria to the tallest
tree.
A new initiative aims to build a
comprehensive tree of life that brings together everything scientists know
about how all species are related, from the tiniest bacteria to the tallest
tree.
Researchers are working to provide the
infrastructure and computational tools to enable automatic updating of the tree
of life, as well as develop the analytical and visualization tools to study it.
Scientists have been building
evolutionary trees for more than 150 years, since Charles Darwin drew the first
sketches in his notebook.
Darwin's theory of evolution explained
that millions of species are related and gave biologists and paleontologists
the enormous challenge of discovering the branching pattern of the tree of
life.
But despite significant progress in
fleshing out the major branches of the tree of life, today there is still no
central place where researchers can go to visualize and analyze the entire tree.
Now, thanks to grants totaling $13
million from the National Science Foundation's (NSF) Assembling, Visualizing,
and Analyzing the Tree of Life (AVAToL) program, three teams of scientists plan
to make that a reality.
"The AVAToL awards are an exciting new
direction for an area that's a foundation of much of biology," says Alan
Townsend, director of NSF's Division of Environmental Biology. "That's
critical to understanding a changing relationship between human society and
Earth's biodiversity."
Figuring out how the millions of species
on Earth are related to one another isn't just important for pinpointing an
antelope's closest kin, or determining if tuna are more closely related to
starfish or hagfish.
Information about evolutionary
relationships is fundamental to comparative biology research. It helps
scientists identify promising new medicines; develop hardier, higher-yielding
crops; and fight infectious diseases such as HIV, anthrax and influenza.
If evolutionary trees are so widely
used, why has assembling them across all life been so hard to achieve?
It's not for lack of research, or data.
Advances in DNA sequencing and evolutionary analysis, discovery of pivotal
early fossils, and novel methods and tools have enabled thousands of new evolutionary
trees to be published in scientific journals each year.
However, most of these focus on
specific, disconnected branches of the tree of life.
Part of the difficulty lies in the sheer
enormity of the task. The largest evolutionary trees to date contain roughly
100,000 groups of organisms.
Assembling the branches for all species
of animals, plants, fungi and microbes--and the countless more still being
named or discovered--will require new computational tools for analyzing large
data sets, for combining diverse kinds of data, and for connecting vast numbers
of published trees into a synthetic whole.
Another difficulty lies in how
scientists typically disseminate their results. A tiny fraction of all
evolutionary trees have been published.
Researchers estimate a mere four percent end up in a database in a
digital form.
Most of the knowledge is locked up in
figures in static journal articles in file formats that may be difficult for
other researchers to download, reanalyze or merge with new information.
What makes this program different from
previous efforts, scientists say, is its scope: its focus on creating an open,
dynamic, evolutionary framework that can be continually refined as new
biodiversity data is collected, and its development of computational and
visualization tools to scale up tree-based evolutionary analyses.
Researchers will be able to go online
and compare their trees to others that have already been published, or download
trees for further study.
They'll also be able to expand the tree,
filling in the missing branches and placing newly named or discovered species
among their relatives.
The goal is to incorporate new trees
automatically, so the complete tree can be continuously updated.
In addition to the creation of an
updatable tree of life, AVAToL scientists will create new tools for the kinds
of research that rely on evolutionary trees and for the collection and analysis
of important evolutionary data, including from fossils critical to the
placement of many branches in the tree of life.
The three NSF-funded AVAToL projects
are:
Automated
and Community-Driven Synthesis of the Tree of Life
Principal Investigator: Karen Cranston,
Duke University and the National Evolutionary Synthesis Center
This project will produce the first
online, comprehensive first-draft tree of all 1.8 million named species,
accessible to both the public and scientists.
Assembly of the tree will incorporate previously published results and
efforts to develop, test and improve methods of data synthesis. This initial
tree of life, called the Open Tree of Life, will not be static. Scientists will
develop tools for researchers to update and revise the tree as new data come
in.
Arbor:
Comparative Analysis Workflows for the Tree of Life
Principal Investigator: Luke Harmon,
University of Idaho
Scientists deal with daunting volumes of
data. One of the most basic challenges
facing researchers is how to organize that information into a usable format
that can inspire new scientific insights. This project team is working to
develop a way to visually portray evolutionary data so scientists can see, at a
glance, how organisms are related. The team will create software tools that
will enable researchers to visualize and analyze data across the tree of life,
enabling research in all areas of comparative biology at multiple evolutionary,
space and time scales. The results have the potential to transform the way
biologists test evolutionary and ecological hypotheses, enabling new research
in fields from medicine to public health, from agriculture to ecology to
genetics.
Next
Generation Phenomics for the Tree of Life
Principal Investigator: Maureen O'Leary,
SUNY-Stony Brook
This team of biologists, computer
scientists and paleontologists will extend and adapt methods from computer
vision, machine learning and natural language processing to enable rapid and
automated study of species' phenotypes on a vast scale across the tree of life.
The team's goal is to develop large phenomic datasets using new methods, and to
provide the scientific community and the public with tools for future such
work. Phenomics is an area of biology that measures the physical and
biochemical traits of organisms as they change in response to genetic mutations
and environmental influences.
Enormous phenomic datasets, many with
images, will foster public interest in biodiversity and the fossil record.
Phenotypic data allow scientists to reconstruct the evolutionary history of
fossil species, in turn crucial for an understanding of the history of life.
This project will leverage recent advances in image analysis and natural
language processing to develop novel approaches to rapidly advance the
collection and analysis of phenotypic data for the tree of life.
-NSF-







No comments:
Post a Comment